Semantic Sholar
-
00:00
1.
GEO2R
播放影片: http://llai.cm.ntu.edu.tw/media/1760
analyze many of the public datasets available in NCBI’s Gene Expression Omnibus or GEO.
You can reach the GEO Datasets page simple by typing “GEO Datasets” in your search engine. I have also included the link in the description below.
http://www.ncbi.nlm.nih.gov/gds
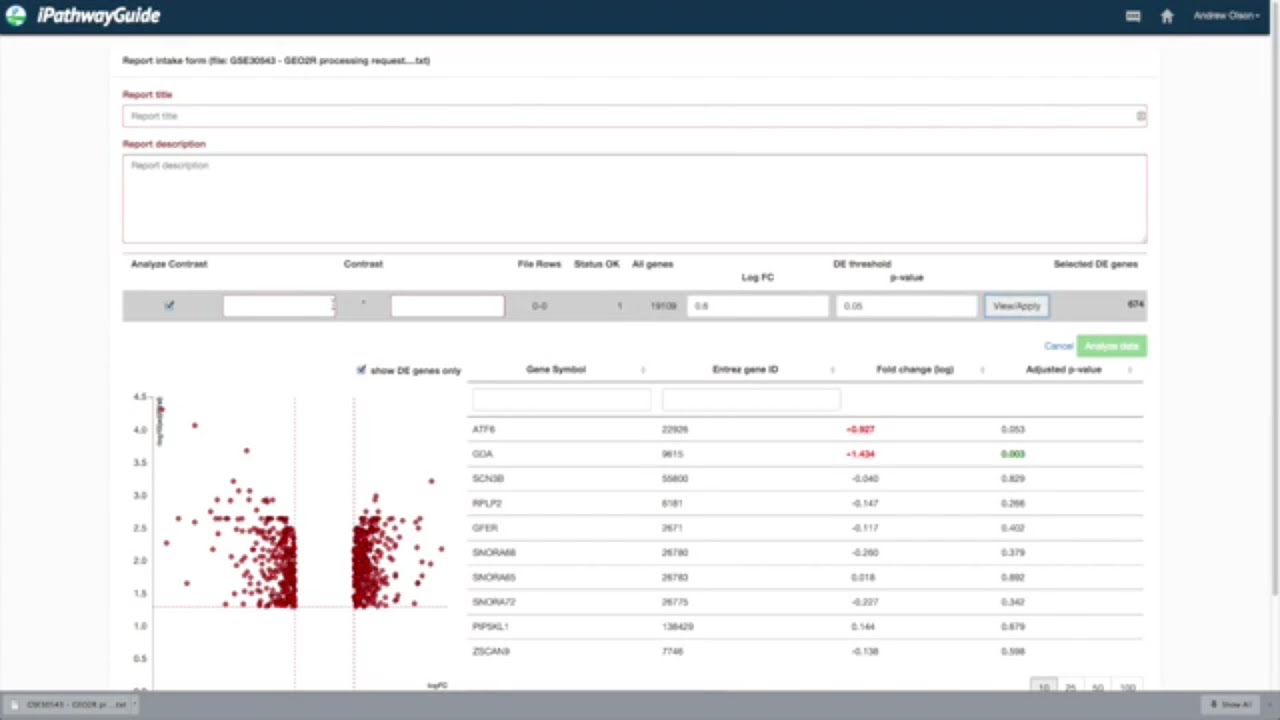
In this example, from the GEO Datasets page, I will search for “breast cancer.” It appears there are over 50,000 breast cancer related datasets in GEO. In this example I’m going to choose this eample looking at the response to TIG1 depletion by clicking on the series number in the description – in this case GSE30543. Here you will find additional information about the experiment. Further down on the page you will see a link that says, “Analyze with GEO2R”
I suggest you open this link in a new tab so you can refer back to the experimental information if needed.
There are additional help tools from within GEO should you need them.
Begin by defining your two groups. In this example we have two groups: TIG1 and the Control.
It is very important when you assign samples to each group that you assign the control group last. Once you have made the proper assignments, scroll down and select “Save all results”.
In about a minute or two you will see a differential expression analysis file.
Once complete, from your browser, choose File, Save Page As to save the file. Be sure to give it a unique identifying name. In this case I will Prepend the name with GSE30543.
You can reach the GEO Datasets page simple by typing “GEO Datasets” in your search engine. I have also included the link in the description below.
http://www.ncbi.nlm.nih.gov/gds
In this example, from the GEO Datasets page, I will search for “breast cancer.” It appears there are over 50,000 breast cancer related datasets in GEO. In this example I’m going to choose this eample looking at the response to TIG1 depletion by clicking on the series number in the description – in this case GSE30543. Here you will find additional information about the experiment. Further down on the page you will see a link that says, “Analyze with GEO2R”
I suggest you open this link in a new tab so you can refer back to the experimental information if needed.
There are additional help tools from within GEO should you need them.
Begin by defining your two groups. In this example we have two groups: TIG1 and the Control.
It is very important when you assign samples to each group that you assign the control group last. Once you have made the proper assignments, scroll down and select “Save all results”.
In about a minute or two you will see a differential expression analysis file.
Once complete, from your browser, choose File, Save Page As to save the file. Be sure to give it a unique identifying name. In this case I will Prepend the name with GSE30543.
- 位置
-
- 資料夾名稱
- 研究基本功
- 發表人
- 賴亮全
- 單位
- 賴亮全教授
- 建立
- 2021-04-06 20:43:48
- 最近修訂
- 2021-04-06 20:45:19
- 長度
- 02:50
- 來源
- https://www.youtube.com/watch?v=nrSAxELqi7Q