維持者: 石忠賢
存放位置: [-20C冰箱][3層][1櫃][Master box盒][27格]
Plasmid datasheet:
DNA sequencing:
存放位置: [-20C冰箱][3層][1櫃][Master box盒][27格]
Plasmid datasheet:
DNA sequencing:
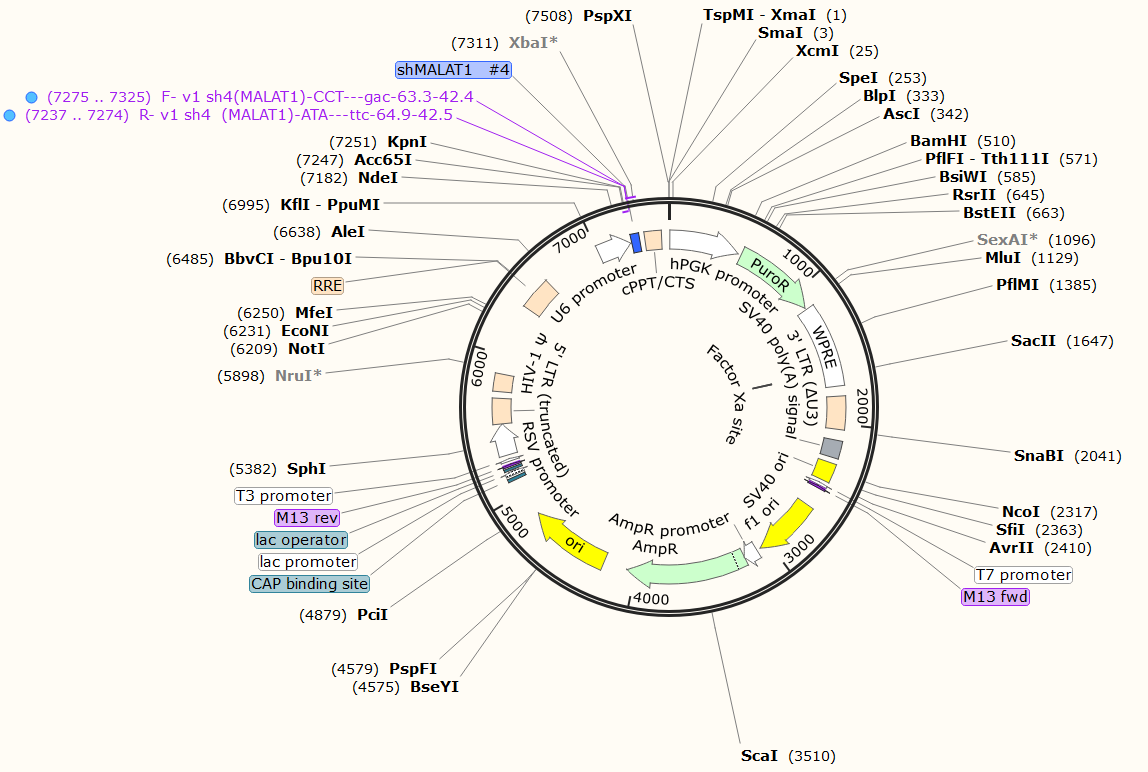
附件:
重點
- 1.SD190716111 of pLKO_shMALAT1 #4 定序結果分析