Comparative Gene-Expression Analysis I: Hybridization-based techniques
-
00:00
1.
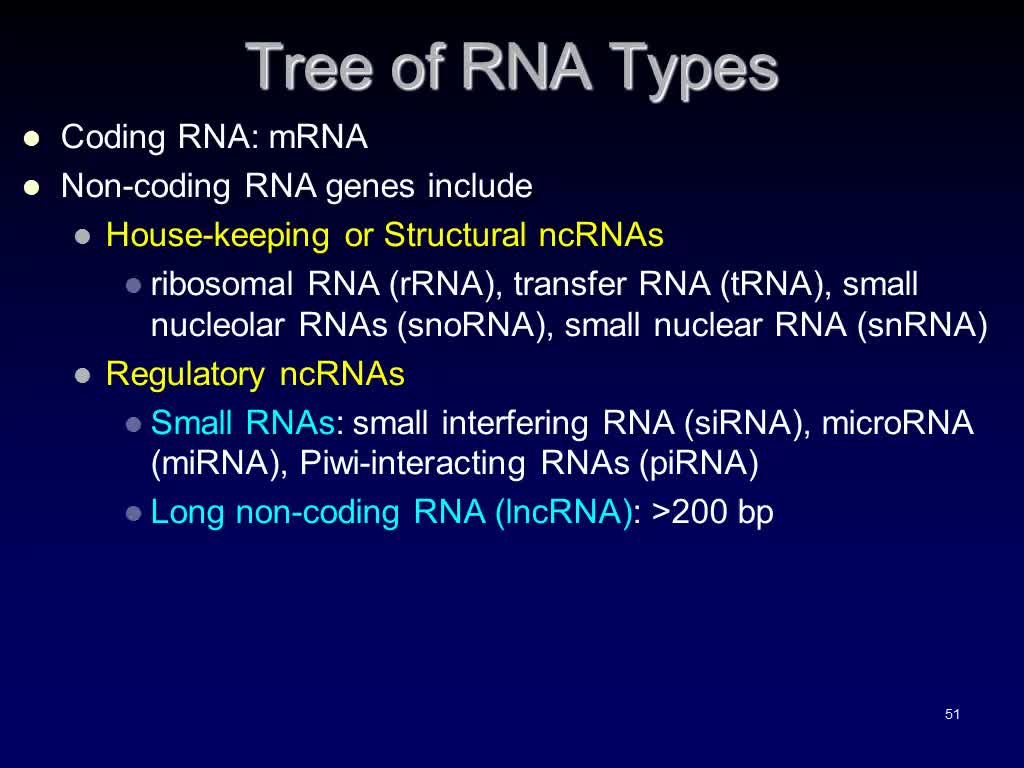
Tree of RNA Types
-
01:29
2.
Non-coding RNA
-
02:15
3.
Major Types of RNA endoribonucleases
-
04:36
4.
Comparative Gene-expression Analysis
-
07:07
5.
Question:Which hybridization technique can be used to detect the amount of polysaccharide?Eastern blottingWestern blottingSouthern blottingNorthern blotting
-
08:18
6.
Agarose gels to resolve large fragments of DNA Polyacrylamide gelsto separate shorter nucleic acids, generally in the range of 1−1000 base pairs, based on the concentration used Gels without a denaturant (e.g., SDS): native gelsSecondary structure affects
-
12:09
7.
Blurry bands Too much DNA (100−250 ng/mm well width) Too much saltBands in the wrong place Heat nucleic acids before running on a native gelRun gel >20 V/cm ( run gel slowly sharper bands)Gel temp. >30 °CLoading buffer floats away Some salts built up i
-
13:07
8.
Question
-
13:48
9.
Hybridization-based techniques1
-
15:43
10.
Northen blotting
-
18:47
11.
Hybridization-based techniques2
-
20:59
12.
Microarray Platforms
-
21:33
13.
Advantages:Favor small sample sizeHigh throughput: gather data on thousands of genes (genome) in a single experimentQuantitative analysisApplication:Gene expressionGenotyping-polymorphisms (SNP) and copy number variationBinding site identification: ChIP-o
-
22:28
14.
High Throughput of Traditional Experiment
-
23:27
15.
Steps for Microarray Experiment
-
25:04
16.
Overview of the stages
-
26:21
17.
Guidelines
-
26:59
18.
Microarray Platforms
-
27:12
19.
In-House Spotted Arrays
-
27:42
20.
microarray animation
-
32:47
21.
URL for Building Microarrayer
-
33:01
22.
picture of arrayer
-
34:03
23.
microarray arrayer video
-
36:01
24.
HEEBO & MEEBO Genome Sets
-
36:22
25.
Microarray Platforms
-
37:14
26.
Prespotted Array Slides
-
39:49
27.
Question
-
39:53
28.
Phosphoamidite Reaction
-
40:52
29.
Feature (Spot) Morphology
-
42:26
30.
QuestionWhy did the samples need to be co-hybridized onto the same slide in two color platform?To reduce reagent costTo minimize microarray numberTo normalize unequal spot sizeTo simplify experimental protocol
-
45:42
31.
Microarray Platforms
-
47:20
32.
Affymetrix
-
47:53
33.
Probe Set and Probe Pair
-
49:02
34.
CheneChip – Affymetrix
-
53:55
35.
Overview of Target preparation
-
54:53
36.
In Vitro Transcription1
-
57:10
37.
In Vitro Transcription2
-
57:30
38.
QuestionIn DNA structure, coding strand (5’ 3’) is also calledWatson strandAnti-sense strandTemplate strandCrick strand
-
58:39
39.
Anti-Sense RNA (Complementary RNA)
-
1:04:55
40.
Overview of target preparation
-
1:05:30
41.
QuestionThe purpose of fragmentation during the process of making cRNA is to reduceContaminationCross-hybridizationAmplification copiesFluorescent intensity
-
1:06:15
42.
Before Hybridization – cRNA Fragmentation1
-
1:07:13
43.
Microarray Platforms
-
1:08:27
44.
Maskless Array Synthesis – Nimblegen
-
1:08:54
45.
Digital Micromirror Device
-
1:09:02
46.
Maskless Array Synthesis
-
1:09:53
47.
QuestionWhat similarities and differences are there in manufacturing microarrays between Affymetrix and NimbleGen?
-
1:10:26
48.
Microarray Platforms
-
1:10:34
49.
BeadChip--Illumina
-
1:11:40
50.
Bead Preparation and Array Production
-
1:13:36
51.
Bead Array: Microwell Fabrication
-
1:13:52
52.
BeadChips
-
1:15:06
53.
Bead Design
-
1:15:55
54.
Decoding Randomly Ordered Bead Arrays
-
1:21:31
55.
Bead Decoding Example: 16 Bead Types
-
1:22:11
56.
BeadChip Products
-
1:23:58
57.
Types of Gene Expression Assays
-
1:25:27
58.
Types of Gene Expression Assays
-
1:28:51
59.
DASL Labeling1
-
1:29:18
60.
DASL Labeling2
-
1:29:44
61.
Whole Genome DASL Workflow
-
1:30:02
62.
QuestionWhat is the major differences of Illumina bead arrays as compared to other platforms?
-
1:30:59
63.
MicroArray Quality Control (MAQC) Project
-
1:31:59
64.
QuestionWhich value of same samples is smaller?Standard deviation (SD)Standard error of the mean (SEM)
-
1:32:34
65.
Coefficient of Variation
-
1:36:21
66.
Repeatability of Expression Signal Within Test Sites
-
1:37:23
67.
Interplatform Data Comparability
-
1:38:25
68.
Scatter Plot
-
1:43:18
69.
Correlation Between Microarray And TaqMan Data
-
1:45:05
70.
QuestionWhich factors will you consider to choose microarray platform? QualityPriceTurn-over rateAvailabilityAll of them
-
1:45:48
71.
Decoding Randomly Ordered Bead Arrays
- 索引
- 筆記
- 討論
- 全螢幕
Comparative Gene-Expression Analysis I: Hybridization-based techniques (0302)
長度: 1:47:36, 瀏覽: 894, 最近修訂: 2021-03-10
-
00:00
1.
Tree of RNA Types
-
01:29
2.
Non-coding RNA
-
02:15
3.
Major Types of RNA endoribonucleases
-
04:36
4.
Comparative Gene-expression Analysis
-
07:07
5.
Question:Which hybridization technique can be used to detect the amount of polysaccharide?Eastern blottingWestern blottingSouthern blottingNorthern blotting
-
08:18
6.
Agarose gels to resolve large fragments of DNA Polyacrylamide gelsto separate shorter nucleic acids, generally in the range of 1−1000 base pairs, based on the concentration used Gels without a denaturant (e.g., SDS): native gelsSecondary structure affects
-
12:09
7.
Blurry bands Too much DNA (100−250 ng/mm well width) Too much saltBands in the wrong place Heat nucleic acids before running on a native gelRun gel >20 V/cm ( run gel slowly sharper bands)Gel temp. >30 °CLoading buffer floats away Some salts built up i
-
13:07
8.
Question
-
13:48
9.
Hybridization-based techniques1
-
15:43
10.
Northen blotting
-
18:47
11.
Hybridization-based techniques2
-
20:59
12.
Microarray Platforms
-
21:33
13.
Advantages:Favor small sample sizeHigh throughput: gather data on thousands of genes (genome) in a single experimentQuantitative analysisApplication:Gene expressionGenotyping-polymorphisms (SNP) and copy number variationBinding site identification: ChIP-o
-
22:28
14.
High Throughput of Traditional Experiment
-
23:27
15.
Steps for Microarray Experiment
-
25:04
16.
Overview of the stages
-
26:21
17.
Guidelines
-
26:59
18.
Microarray Platforms
-
27:12
19.
In-House Spotted Arrays
-
27:42
20.
microarray animation
-
32:47
21.
URL for Building Microarrayer
-
33:01
22.
picture of arrayer
-
34:03
23.
microarray arrayer video
-
36:01
24.
HEEBO & MEEBO Genome Sets
-
36:22
25.
Microarray Platforms
-
37:14
26.
Prespotted Array Slides
-
39:49
27.
Question
-
39:53
28.
Phosphoamidite Reaction
-
40:52
29.
Feature (Spot) Morphology
-
42:26
30.
QuestionWhy did the samples need to be co-hybridized onto the same slide in two color platform?To reduce reagent costTo minimize microarray numberTo normalize unequal spot sizeTo simplify experimental protocol
-
45:42
31.
Microarray Platforms
-
47:20
32.
Affymetrix
-
47:53
33.
Probe Set and Probe Pair
-
49:02
34.
CheneChip – Affymetrix
-
53:55
35.
Overview of Target preparation
-
54:53
36.
In Vitro Transcription1
-
57:10
37.
In Vitro Transcription2
-
57:30
38.
QuestionIn DNA structure, coding strand (5’ 3’) is also calledWatson strandAnti-sense strandTemplate strandCrick strand
-
58:39
39.
Anti-Sense RNA (Complementary RNA)
-
1:04:55
40.
Overview of target preparation
-
1:05:30
41.
QuestionThe purpose of fragmentation during the process of making cRNA is to reduceContaminationCross-hybridizationAmplification copiesFluorescent intensity
-
1:06:15
42.
Before Hybridization – cRNA Fragmentation1
-
1:07:13
43.
Microarray Platforms
-
1:08:27
44.
Maskless Array Synthesis – Nimblegen
-
1:08:54
45.
Digital Micromirror Device
-
1:09:02
46.
Maskless Array Synthesis
-
1:09:53
47.
QuestionWhat similarities and differences are there in manufacturing microarrays between Affymetrix and NimbleGen?
-
1:10:26
48.
Microarray Platforms
-
1:10:34
49.
BeadChip--Illumina
-
1:11:40
50.
Bead Preparation and Array Production
-
1:13:36
51.
Bead Array: Microwell Fabrication
-
1:13:52
52.
BeadChips
-
1:15:06
53.
Bead Design
-
1:15:55
54.
Decoding Randomly Ordered Bead Arrays
-
1:21:31
55.
Bead Decoding Example: 16 Bead Types
-
1:22:11
56.
BeadChip Products
-
1:23:58
57.
Types of Gene Expression Assays
-
1:25:27
58.
Types of Gene Expression Assays
-
1:28:51
59.
DASL Labeling1
-
1:29:18
60.
DASL Labeling2
-
1:29:44
61.
Whole Genome DASL Workflow
-
1:30:02
62.
QuestionWhat is the major differences of Illumina bead arrays as compared to other platforms?
-
1:30:59
63.
MicroArray Quality Control (MAQC) Project
-
1:31:59
64.
QuestionWhich value of same samples is smaller?Standard deviation (SD)Standard error of the mean (SEM)
-
1:32:34
65.
Coefficient of Variation
-
1:36:21
66.
Repeatability of Expression Signal Within Test Sites
-
1:37:23
67.
Interplatform Data Comparability
-
1:38:25
68.
Scatter Plot
-
1:43:18
69.
Correlation Between Microarray And TaqMan Data
-
1:45:05
70.
QuestionWhich factors will you consider to choose microarray platform? QualityPriceTurn-over rateAvailabilityAll of them
-
1:45:48
71.
Decoding Randomly Ordered Bead Arrays
- 位置
-
- 資料夾名稱
- 2021
- 發表人
- 賴亮全
- 單位
- 賴亮全教授
- 建立
- 2021-03-02 17:28:34
- 最近修訂
- 2021-03-10 12:41:57
- 長度
- 1:47:36