Comparative Gene-Expression Analysis I: Hybridization-based techniques
-
00:00
1.
Review
-
01:33
2.
Gene Ontology
-
04:58
3.
Pathway Analysis
-
08:54
4.
Ingenuity Pathway Analysis (IPA)
-
12:10
5.
Pathway Analysis
-
15:21
6.
MetaCore
-
15:41
7.
GeneMANIA
-
16:45
8.
Gene Set Enrichment Analysis
-
24:09
9.
Gene Set Enrichment Analysis
-
25:29
10.
MSigDB Collections
-
26:34
11.
Pathway Analysis
-
27:43
12.
Color Objects in KEGG Pathways
-
28:19
13.
BIOCARTA Pathway
-
28:43
14.
The Database for Annotation, Visualization and Integrated Discovery (DAVID)
-
29:37
15.
Reference Databases in DAVID
-
30:27
16.
Transcription Factor Motif Search1
-
33:36
17.
Ways to describe DNA motif
-
39:24
18.
Transcription Factor Motif Search3
-
42:02
19.
Direct TFM search
-
44:15
20.
Position weight matrix method
-
45:04
21.
Greedy Algorithm
-
47:32
22.
C. Phylogenetic footprinting methodUse orthologous promoter sequences of a single gene from multiple speciese.g., ConSite, Phylonet, Phylo-ScanProblems:If the species are too closely related, seq. alignment is obvious but uninformativeIf the species are
-
48:39
23.
MOTIF Similarity Detection Tool
-
49:17
24.
example
-
50:38
25.
Databases
-
51:08
26.
Nucleotide Sequences
-
51:46
27.
Gene Expression
-
52:49
28.
GEO Structure
-
55:34
29.
GEO2R
-
56:05
30.
Transcription Factor Binding Site
-
56:19
31.
Genome Browsers
-
57:12
32.
UCSC Genome Browser
-
58:16
33.
Multipurpose Portals
-
58:35
34.
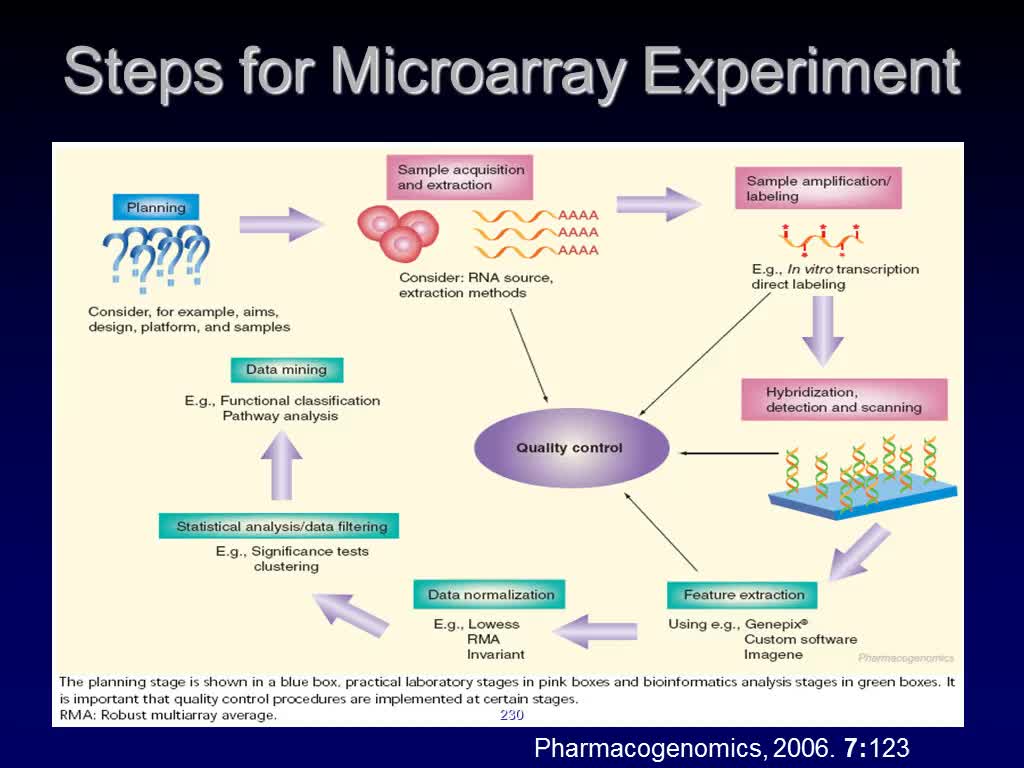
Workflow of Array Analysis
-
59:29
35.
Array Analysis1
-
59:56
36.
Array Analysis2
-
1:00:19
37.
Array Analysis3
-
1:00:45
38.
Array Analysis4
-
1:00:47
39.
Array Analysis3
-
1:01:01
40.
Array Analysis4
-
1:01:22
41.
Array Analysis5
- 索引
- 筆記
- 討論
- 全螢幕
Comparative Gene-Expression Analysis I: Hybridization-based techniques (0323)
長度: 1:02:03, 瀏覽: 820, 最近修訂: 2021-03-23
-
00:00
1.
Review
-
01:33
2.
Gene Ontology
-
04:58
3.
Pathway Analysis
-
08:54
4.
Ingenuity Pathway Analysis (IPA)
-
12:10
5.
Pathway Analysis
-
15:21
6.
MetaCore
-
15:41
7.
GeneMANIA
-
16:45
8.
Gene Set Enrichment Analysis
-
24:09
9.
Gene Set Enrichment Analysis
-
25:29
10.
MSigDB Collections
-
26:34
11.
Pathway Analysis
-
27:43
12.
Color Objects in KEGG Pathways
-
28:19
13.
BIOCARTA Pathway
-
28:43
14.
The Database for Annotation, Visualization and Integrated Discovery (DAVID)
-
29:37
15.
Reference Databases in DAVID
-
30:27
16.
Transcription Factor Motif Search1
-
33:36
17.
Ways to describe DNA motif
-
39:24
18.
Transcription Factor Motif Search3
-
42:02
19.
Direct TFM search
-
44:15
20.
Position weight matrix method
-
45:04
21.
Greedy Algorithm
-
47:32
22.
C. Phylogenetic footprinting methodUse orthologous promoter sequences of a single gene from multiple speciese.g., ConSite, Phylonet, Phylo-ScanProblems:If the species are too closely related, seq. alignment is obvious but uninformativeIf the species are
-
48:39
23.
MOTIF Similarity Detection Tool
-
49:17
24.
example
-
50:38
25.
Databases
-
51:08
26.
Nucleotide Sequences
-
51:46
27.
Gene Expression
-
52:49
28.
GEO Structure
-
55:34
29.
GEO2R
-
56:05
30.
Transcription Factor Binding Site
-
56:19
31.
Genome Browsers
-
57:12
32.
UCSC Genome Browser
-
58:16
33.
Multipurpose Portals
-
58:35
34.
Workflow of Array Analysis
-
59:29
35.
Array Analysis1
-
59:56
36.
Array Analysis2
-
1:00:19
37.
Array Analysis3
-
1:00:45
38.
Array Analysis4
-
1:00:47
39.
Array Analysis3
-
1:01:01
40.
Array Analysis4
-
1:01:22
41.
Array Analysis5
- 位置
-
- 資料夾名稱
- 2021
- 發表人
- 賴亮全
- 單位
- 賴亮全教授
- 建立
- 2021-03-23 16:20:25
- 最近修訂
- 2021-03-23 18:08:25
- 長度
- 1:02:03