Comparative Gene-Expression Analysis I: Hybridization-based techniques
-
00:00
1.
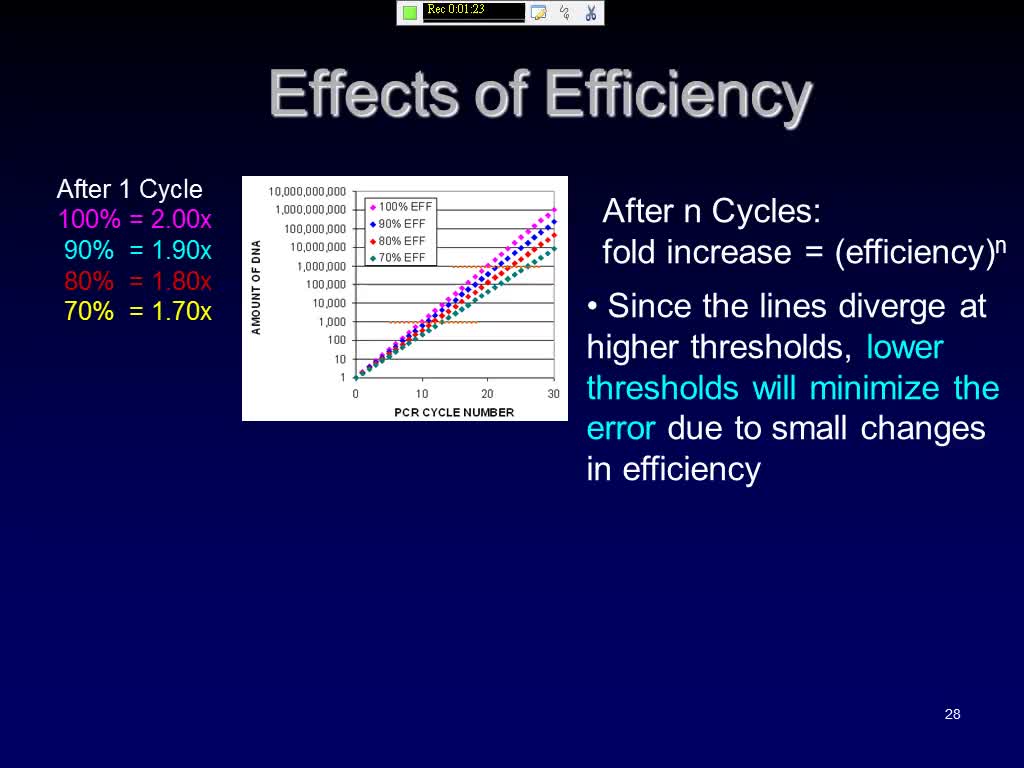
Effects of Efficiency
-
01:49
2.
Melting Curve
-
03:27
3.
Melting Curve
-
04:23
4.
Relative Quantity Value: 2-ΔΔCT
-
09:57
5.
Question:Given the following Ct values, please calculate the fold change of YFG in experiment group.
-
10:44
6.
Real Time - PCR
-
11:57
7.
SYBR Green Dye Assay
-
12:40
8.
Fluorescence Resonance Energy Transfer (FRET)
-
13:58
9.
Principle of TaqMan Assay
-
16:33
10.
TaqMan RT-PCR assay
-
18:48
11.
LightCycler Probes
-
19:51
12.
Molecular Beacons
-
20:30
13.
QuestionHow to amplify strand-specific DNA by PCR?
-
21:07
14.
Strand Specific PCR
-
24:45
15.
Absolute vs. Relative Quantification for qPCR
-
25:51
16.
QuestionWhat is the difference between analog vs digital signal?
-
26:42
17.
Digital PCR
-
31:46
18.
Digital PCR
- 索引
- 筆記
- 討論
- 全螢幕
Comparative Gene-Expression Analysis II: PCR-based techniques (0330)
長度: 33:51, 瀏覽: 777, 最近修訂: 2021-03-30
-
00:00
1.
Effects of Efficiency
-
01:49
2.
Melting Curve
-
03:27
3.
Melting Curve
-
04:23
4.
Relative Quantity Value: 2-ΔΔCT
-
09:57
5.
Question:Given the following Ct values, please calculate the fold change of YFG in experiment group.
-
10:44
6.
Real Time - PCR
-
11:57
7.
SYBR Green Dye Assay
-
12:40
8.
Fluorescence Resonance Energy Transfer (FRET)
-
13:58
9.
Principle of TaqMan Assay
-
16:33
10.
TaqMan RT-PCR assay
-
18:48
11.
LightCycler Probes
-
19:51
12.
Molecular Beacons
-
20:30
13.
QuestionHow to amplify strand-specific DNA by PCR?
-
21:07
14.
Strand Specific PCR
-
24:45
15.
Absolute vs. Relative Quantification for qPCR
-
25:51
16.
QuestionWhat is the difference between analog vs digital signal?
-
26:42
17.
Digital PCR
-
31:46
18.
Digital PCR
- 位置
-
- 資料夾名稱
- 2021
- 發表人
- 賴亮全
- 單位
- 賴亮全教授
- 建立
- 2021-03-30 16:17:46
- 最近修訂
- 2021-03-30 20:38:33
- 長度
- 33:51