Comparative Gene-Expression Analysis I: Hybridization-based techniques
-
00:00
1.
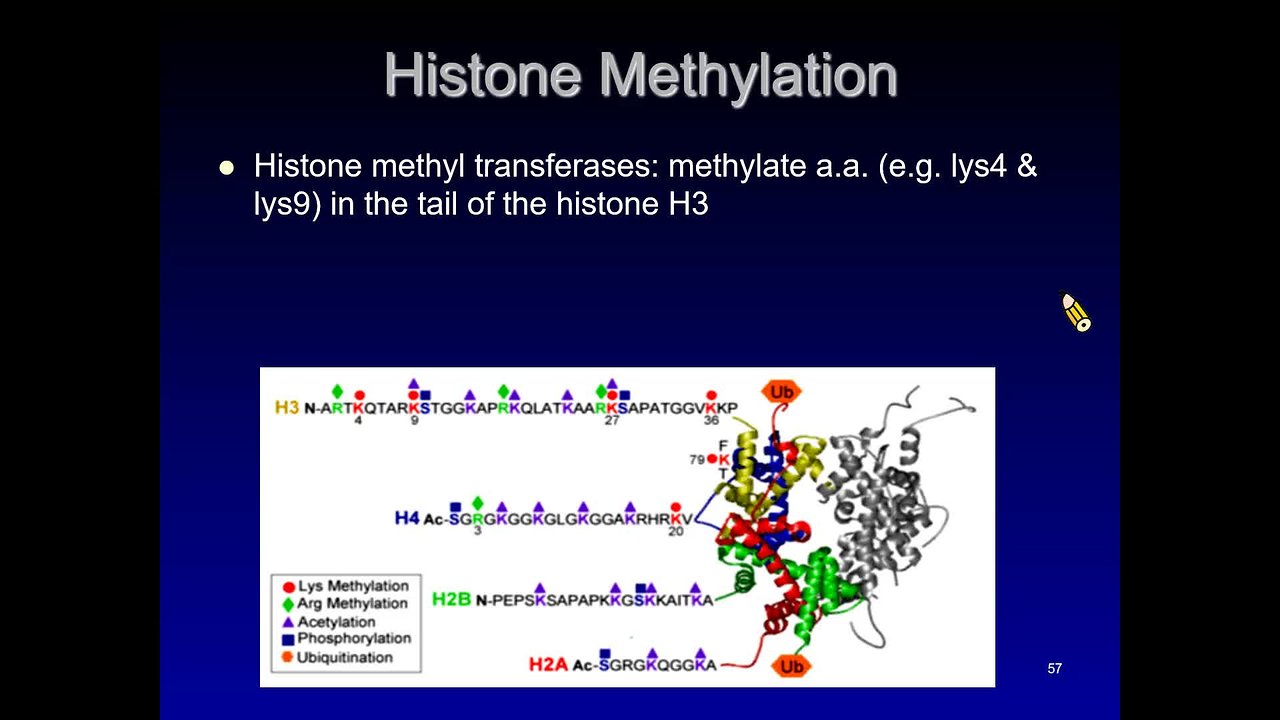
Histone Mehtylation
-
01:01
2.
Histone Modification and Histone Codes
-
01:51
3.
Histone Acetylation
-
03:54
4.
Aberrant Histone Modifications in Cancer
-
05:26
5.
Locus-Specific Genetic Changes
-
07:24
6.
Locus-Specific Epigenetic Changes
-
08:51
7.
Epigenetic Mechanisms
-
09:08
8.
Polycomb–Trithorax Protein Complexes
-
09:55
9.
ATP-Based Chromatin-Remodeling Complexes
-
10:38
10.
Epigenetic Mechanisms
-
10:43
11.
PRION
-
11:54
12.
期末報告—互評
-
13:11
13.
Epigenetic Mechanisms
-
13:24
14.
Non-coding RNA
-
14:48
15.
Non-coding RNA classification
-
15:19
16.
Function of Non-coding RNA
-
16:16
17.
Number of HGNC Gene Symbols
-
16:46
18.
Nomenclature of rRNA
-
18:31
19.
Nomenclature of rRNA
-
19:47
20.
Nomenclature of tRNA
-
21:00
21.
Panoramic RNA display by overcoming RNA modification aborted sequencingA combinatorial enzymatic treatment to remove key RNA modifications AlkB enzyme: prevent blockage of reverse transcriptionT4PNK enzyme: prevent blockage of adapter ligation
-
22:24
22.
Systematically discovery of LncRNA
-
24:33
23.
Origins of LncRNAs
-
25:33
24.
Long Non-coding RNA Nomenclature
-
27:13
25.
Long Non-coding RNA Nomenclature
-
28:30
26.
LncRNA Functions
-
31:04
27.
Long non-coding RNAs as a Source of New Peptides
-
33:32
28.
Tools to predict translation potential of noncoding RNA
-
34:11
29.
dUTP Based Strand-Specific Sequencing
-
35:45
30.
dUTP Based Strand-Specific Sequencing
-
36:41
31.
Methods for Studying lncRNAs
-
38:45
32.
Methods for Detecting Protein-RNA Interactions
-
39:18
33.
RNase Protection Assay
-
40:18
34.
Electrophoretic Mobility Shift Assay (EMSA)
-
42:40
35.
RIP-Seq
-
43:07
36.
Question
-
43:26
37.
Protein A, G and L Antibody-Binding Ligands
-
45:18
38.
CLIP-Seq
-
45:38
39.
Crosslink Reagents
-
46:41
40.
RNA Pulldown Assay
-
47:49
41.
Aptamer
-
50:03
42.
RNA Fluorescent In-Situ Hybridiation (FISH)
-
50:49
43.
Prediction of Human lncRNA-Protein Interactions
-
51:25
44.
ENCORI
-
52:06
45.
RNA-Protein Interaction Prediction (RPISeq)
-
52:58
46.
Methods for Studying lncRNAs
-
54:49
47.
Summary graph
-
55:50
48.
Summary Table
-
56:07
49.
Methods for Studying lncRNAs
-
56:25
50.
Circular RNA
-
57:27
51.
Circular RNA
-
58:55
52.
Circular RNA
-
1:00:20
53.
Mechanisms of circRNA translation
-
1:01:44
54.
Functions of circRNA
-
1:03:47
55.
circRNA-Protein Interactions
-
1:04:15
56.
Circular RNA Interactome
-
1:04:24
57.
circRNA databases
-
1:04:30
58.
Post-Transcriptional Regulation
-
1:05:41
59.
microRNA1
-
1:06:48
60.
microRNA2
-
1:07:34
61.
microRNA3
-
1:08:50
62.
Biogenesis of microRNA
-
1:12:50
63.
miRNA Nomenclature
-
1:15:37
64.
Functions of miRNA
-
1:15:59
65.
How to research miRNA?
-
1:16:12
66.
Approaches to miRNA Discovery1
-
1:17:22
67.
Approaches to miRNA Discovery2
-
1:18:54
68.
Approaches to miRNA Discovery3
-
1:19:15
69.
Alternative Reverse Transcription Methodologies to Generate cDNA from miRNA
-
1:19:59
70.
cDNA synthesis by using stem-loop and linear miRNA-specific primers
-
1:20:52
71.
RT-PCR miRNA Assays
-
1:21:57
72.
Approaches to miRNA Discovery4
-
1:22:30
73.
Detection of microRNAs by In Situ Hybridization (ISH)
-
1:23:44
74.
Approaches to miRNA Discovery5
-
1:24:42
75.
How to research miRNA?
-
1:24:49
76.
Approaches to Predict miRNA Target
-
1:26:01
77.
List of Tools for miRNA Target Prediction
-
1:26:20
78.
Question
-
1:26:29
79.
miRSystem
-
1:28:14
80.
Luciferase Reporter Assay for MiRNA Target
-
1:29:29
81.
Analyses of miRNA Targets Using NGS
-
1:29:51
82.
How to research miRNA?
-
1:29:58
83.
Nanostring nCounter miRNA Assay
-
1:30:54
84.
AntimiRs
-
1:31:16
85.
Modified Nucleic Acid
-
1:32:11
86.
AntimiRs
-
1:33:04
87.
miR mimics
-
1:33:53
88.
Tools4miRs
-
1:34:13
89.
Competing Endogenous RNA (ceRNA)
-
1:35:35
90.
Discovery of RNA interference
-
1:36:06
91.
Silencing of unc-22 gene in C. elegans
-
1:36:54
92.
Short Interfering RNA (siRNA)
-
1:38:27
93.
Mechanism of siRNA
-
1:39:23
94.
General Design Guidelines for siRNA
-
1:41:14
95.
Off-Target Effects
-
1:42:17
96.
Appropriate Experimental Controls to Minimize Off-Target Effects (OTEs)
-
1:43:55
97.
siRNA Expression
-
1:45:51
98.
siRNA Delivery & Processing
-
1:47:29
99.
siRNA as Human Therapeutics
-
1:48:01
100.
Differences Between siRNA & miRNA
-
1:49:30
101.
Difference Between miRNA & siRNA
-
1:50:13
102.
Morpholino
-
1:50:59
103.
Summary
-
1:51:55
104.
Question
-
1:52:36
105.
Question
-
1:52:46
106.
Small Activating RNAs (saRNAs)
-
00:00
1.
Histone Mehtylation
-
01:01
2.
Histone Modification and Histone Codes
-
01:51
3.
Histone Acetylation
-
03:54
4.
Aberrant Histone Modifications in Cancer
-
05:26
5.
Locus-Specific Genetic Changes
-
07:24
6.
Locus-Specific Epigenetic Changes
-
08:51
7.
Epigenetic Mechanisms
-
09:08
8.
Polycomb–Trithorax Protein Complexes
-
09:55
9.
ATP-Based Chromatin-Remodeling Complexes
-
10:38
10.
Epigenetic Mechanisms
-
10:43
11.
PRION
-
11:54
12.
期末報告—互評
-
13:11
13.
Epigenetic Mechanisms
-
13:24
14.
Non-coding RNA
-
14:48
15.
Non-coding RNA classification
-
15:19
16.
Function of Non-coding RNA
-
16:16
17.
Number of HGNC Gene Symbols
-
16:46
18.
Nomenclature of rRNA
-
18:31
19.
Nomenclature of rRNA
-
19:47
20.
Nomenclature of tRNA
-
21:00
21.
Panoramic RNA display by overcoming RNA modification aborted sequencingA combinatorial enzymatic treatment to remove key RNA modifications AlkB enzyme: prevent blockage of reverse transcriptionT4PNK enzyme: prevent blockage of adapter ligation
-
22:24
22.
Systematically discovery of LncRNA
-
24:33
23.
Origins of LncRNAs
-
25:33
24.
Long Non-coding RNA Nomenclature
-
27:13
25.
Long Non-coding RNA Nomenclature
-
28:30
26.
LncRNA Functions
-
31:04
27.
Long non-coding RNAs as a Source of New Peptides
-
33:32
28.
Tools to predict translation potential of noncoding RNA
-
34:11
29.
dUTP Based Strand-Specific Sequencing
-
35:45
30.
dUTP Based Strand-Specific Sequencing
-
36:41
31.
Methods for Studying lncRNAs
-
38:45
32.
Methods for Detecting Protein-RNA Interactions
-
39:18
33.
RNase Protection Assay
-
40:18
34.
Electrophoretic Mobility Shift Assay (EMSA)
-
42:40
35.
RIP-Seq
-
43:07
36.
Question
-
43:26
37.
Protein A, G and L Antibody-Binding Ligands
-
45:18
38.
CLIP-Seq
-
45:38
39.
Crosslink Reagents
-
46:41
40.
RNA Pulldown Assay
-
47:49
41.
Aptamer
-
50:03
42.
RNA Fluorescent In-Situ Hybridiation (FISH)
-
50:49
43.
Prediction of Human lncRNA-Protein Interactions
-
51:25
44.
ENCORI
-
52:06
45.
RNA-Protein Interaction Prediction (RPISeq)
-
52:58
46.
Methods for Studying lncRNAs
-
54:49
47.
Summary graph
-
55:50
48.
Summary Table
-
56:07
49.
Methods for Studying lncRNAs
-
56:25
50.
Circular RNA
-
57:27
51.
Circular RNA
-
58:55
52.
Circular RNA
-
1:00:20
53.
Mechanisms of circRNA translation
-
1:01:44
54.
Functions of circRNA
-
1:03:47
55.
circRNA-Protein Interactions
-
1:04:15
56.
Circular RNA Interactome
-
1:04:24
57.
circRNA databases
-
1:04:30
58.
Post-Transcriptional Regulation
-
1:05:41
59.
microRNA1
-
1:06:48
60.
microRNA2
-
1:07:34
61.
microRNA3
-
1:08:50
62.
Biogenesis of microRNA
-
1:12:50
63.
miRNA Nomenclature
-
1:15:37
64.
Functions of miRNA
-
1:15:59
65.
How to research miRNA?
-
1:16:12
66.
Approaches to miRNA Discovery1
-
1:17:22
67.
Approaches to miRNA Discovery2
-
1:18:54
68.
Approaches to miRNA Discovery3
-
1:19:15
69.
Alternative Reverse Transcription Methodologies to Generate cDNA from miRNA
-
1:19:59
70.
cDNA synthesis by using stem-loop and linear miRNA-specific primers
-
1:20:52
71.
RT-PCR miRNA Assays
-
1:21:57
72.
Approaches to miRNA Discovery4
-
1:22:30
73.
Detection of microRNAs by In Situ Hybridization (ISH)
-
1:23:44
74.
Approaches to miRNA Discovery5
-
1:24:42
75.
How to research miRNA?
-
1:24:49
76.
Approaches to Predict miRNA Target
-
1:26:01
77.
List of Tools for miRNA Target Prediction
-
1:26:20
78.
Question
-
1:26:29
79.
miRSystem
-
1:28:14
80.
Luciferase Reporter Assay for MiRNA Target
-
1:29:29
81.
Analyses of miRNA Targets Using NGS
-
1:29:51
82.
How to research miRNA?
-
1:29:58
83.
Nanostring nCounter miRNA Assay
-
1:30:54
84.
AntimiRs
-
1:31:16
85.
Modified Nucleic Acid
-
1:32:11
86.
AntimiRs
-
1:33:04
87.
miR mimics
-
1:33:53
88.
Tools4miRs
-
1:34:13
89.
Competing Endogenous RNA (ceRNA)
-
1:35:35
90.
Discovery of RNA interference
-
1:36:06
91.
Silencing of unc-22 gene in C. elegans
-
1:36:54
92.
Short Interfering RNA (siRNA)
-
1:38:27
93.
Mechanism of siRNA
-
1:39:23
94.
General Design Guidelines for siRNA
-
1:41:14
95.
Off-Target Effects
-
1:42:17
96.
Appropriate Experimental Controls to Minimize Off-Target Effects (OTEs)
-
1:43:55
97.
siRNA Expression
-
1:45:51
98.
siRNA Delivery & Processing
-
1:47:29
99.
siRNA as Human Therapeutics
-
1:48:01
100.
Differences Between siRNA & miRNA
-
1:49:30
101.
Difference Between miRNA & siRNA
-
1:50:13
102.
Morpholino
-
1:50:59
103.
Summary
-
1:51:55
104.
Question
-
1:52:36
105.
Question
-
1:52:46
106.
Small Activating RNAs (saRNAs)
- 位置
-
- 資料夾名稱
- 2021
- 發表人
- 賴亮全
- 單位
- 賴亮全教授
- 建立
- 2021-05-18 14:11:47
- 最近修訂
- 2021-05-18 19:37:13
- 長度
- 1:53:34